
Professor Michelle Peckham
- Position: Professor
- Areas of expertise: Cytoskeleton; actin; myosin; molecular motors; myosin; kinesin; Microscopy; super-resolution imaging; electron microscopy; biochemical techniques
- Email: M.Peckham@leeds.ac.uk
- Phone: +44(0)113 343 4348
- Location: 8.106 Astbury
- Website: Contractility Group | X | LinkedIn | Googlescholar | Researchgate | ORCID
Profile
I graduated in Biology (Physiology of Organisms) at the University of York in 1981, and went on to do my PhD with Prof. Roger Woledge in the Physiology Department at University College London, on the energetics of muscle contraction. I was awarded my PhD in 1984, worked briefly with Roger for a couple of months, before moving to the Biophysics Department at Kings College London, where I worked with Professor Malcolm Irving to demonstrate that birefringence could be used as a reporter of myosin cross bridge orientation in skeletal muscle (1985-7). I then worked at UCSF (San Francisco)(1987-8) before returning to the Biology Department at the University of York, to work with Professors David White and John Sparrow on insect flight muscle kinetics (1988-1990). As part of that work, we used the flight muscle of Drosophila Melanogaster as a model system. Mutations in contractile proteins were engineered into flight muscle actin, and I used sophisticated biophysical measurements to determine effects on contraction, resulting in a paper in Nature (1990). I was awarded a Royal Society University Research Fellowship to move to the Biophysics Department, KCL in 1990, where I started my own lab, and began to use molecular biology and cell culture techniques to investigate the cytoskeleton in cultured muscle cells. I moved to the University of Leeds in 1997, becoming a Lecturer, Senior Lecturer, Reader then Professor in 2010. I developed a strong interest in imaging and confocal microscopy alongside my interests in the cytoskeleton, developing super-resolution approaches at Leeds about 5-10 years ago, including the development of Affimers for STORM. My interests have also expanded out to more structural biology approaches, including the discovery and characterisation of single alpha helices (SAH) domains in proteins and most recently my laboratory solved the structure of the shutdown state of myosin using Cryo-EM. I have trained over 40 postgraduate and postdoctoral researchers. I was the president of the Royal Microscopical Society (RMS) from 2016-2019 and am currently the Executive Honorary Secretary of the RMS. I am currently a Wellcome Trust Investigator, working on how the activity of motor proteins is regulated.
Responsibilities
- Executive Honorary Secretary, Royal Microscopical Society
Research interests
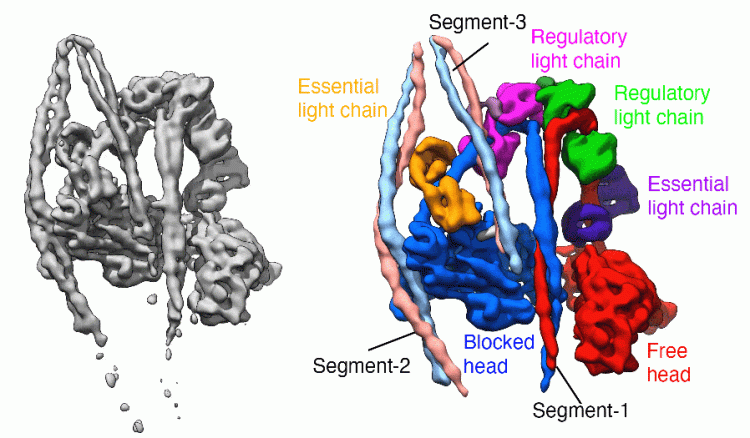
Our laboratory has broad interests in the cytoskeleton, from basic research into how myosins and other motor proteins perform their functions in cells, to how mutations in cytoskeletal proteins cause disease. We use a wide range of approaches to uncover how myosins work, from solving the structure by Cryo-EM to imaging myosin and the cytoskeleton within cells using confocal, super-resolution and electron microscopy, and a range of biochemical and biophysical approaches.
We have solved the structure of the shutdown state of smooth muscle myosin (Scarff et al. December 2020, Nature) as shown above, using CryoEM. This reveals for the first time how myosin adopts the shutdown state, and how phosphorylation of the regulatory light chain activates the molecule.
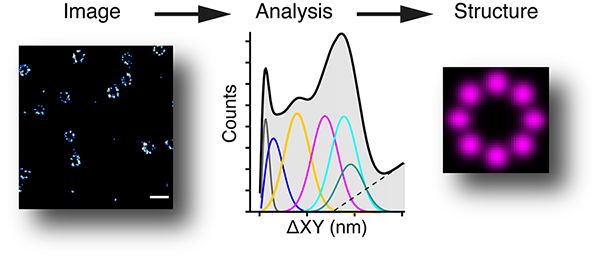
We have a very strong interest in imaging, including super-resolution imaging. We have built a 3D PALM/STORM system, that allows a resolution of ~10nm (about 20 times better than a normal wide-field microscope), and an iSIM (instant structured illumination microscope) which is very good for fast live cell imaging. The image below shows a 3D STORM of nuclear pores (from Jonas Ries’s lab) and how we can analyse them to find the underlying structure (Curd et al., Nanoletters, 2020).
We are collaborating with Darren Tomlinson's group to raise small non-antibody binding proteins (~12kDa in size) called Affimers to cytoskeletal proteins, to improve our super-resolution imaging, and have also a range of Affimers to different proteins including tubulin (recently published in E-life: Tiede et al., 2017; Lopata et al., Sci. Reports 2018). We have also used STED (stimulated emission depletion microscopy) to show how Z-discs, labelled with Affimers to specific proteins, are altered in diseased (dilated cardiomyopathy) hearts (Parker et al., Frontiers in Cardiovascular Medicine 2023). You can find out more about Affimers and their use in imaging in our recent review (Cordell et al., 2022, Journal of Cell Science).
We work on the roles of myosins in diseases such as cancer (Makowska et al., Cell Reports, 2015), where we find that specific myosins are overexpressed and contribute to the phenotype of prostate cancer.
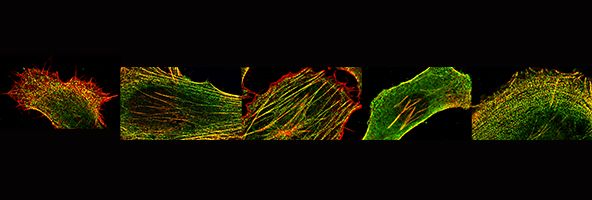
We are also interested in how muscle cells differentiate in culture - and how they can organise the cytoskeleton into beautifully regular structures, as show here for a cultured human skeletal myotube, stained for skeletal myosin (green), and use this as a model system to investigate the effects of mutations in muscle myosins (Parker et al., Journal of Molecular Biology, 2018)

PhD students in the lab are also studying aspects of these problems, including using AI to analyse super-resolution microscopy images (EPSPRC AI CDT funded), deriving Affimers to visualise smooth and non-muscle myosins in cells (BBSRC DTP), and investigating the effect of heart muscle diseases on the organisation of sarcomeric and membrane proteins (BHF 4 year BHF programme).
The Astbury Centre for Structural Molecular Biology
<h4>Research projects</h4> <p>Some research projects I'm currently working on, or have worked on, will be listed below. Our list of all <a href="https://biologicalsciences.leeds.ac.uk/dir/research-projects">research projects</a> allows you to view and search the full list of projects in the faculty.</p>Primary investigator (PI)
- "NanoRAM: Emerging Nanotools for Soft Matter Characterisation and Manipulation"
- Developing novel tools to target the cytoskeleton in health and disease: a UK-Australia collaboration
- Regulatory mechanisms in myosins
- Super-resolution imaging across the Biosciences
- Understanding how proteins are organised in the Z-disk, a super-resolution approach
- Understanding the mechanisms underlying myosinopathies
Co-investigator (Co-I)
- Dissecting SOS1 and SOS2 isoform-specific regulation of Ras signalling through biochemical, structural, and Affimer-based approaches
- Imaging the interactome
Qualifications
- PhD, London (1984)
- BA, York
Professional memberships
- Fellow of the Royal Microscopical Society
- Member of British Society for Molecular Biology
- Fellow of the Royal Society for Biology
Student education
Undergraduate project topics:
- Imaging, Microscopy, cytoskeleton, diseases linked to cytoskeletal proteins (including proteins in the muscle cytoskeleton)
Postgraduate studentship areas:
- PhD opportunities in the cytoskeleton, roles of cytoskeletal proteins in disease - including cardiac myosin heavy chain mutations that cause heart disease, mutations in non-muscle myosin 2A that causes bleeding disorders, mutations in myosins related to other diseases such as deafness and blindness. Role of the cytoskeleton in cancer. Super-resolution imaging. CryoEM and structure determination.
See also:
- Faculty Graduate School
- FindaPhD Project details:
Research groups and institutes
- Cell and Organismal Biology
- Structural Biology
- Cancer and cell signalling
- Protein structure, stability and dynamics
- Infectious diseases
Projects
-
<li><a href="//phd.leeds.ac.uk/project/1928-how-does-myosin-10-contribute-to-filopodia-formation-and-stability?">How does myosin 10 contribute to filopodia formation and stability?</a></li>

