Biomolecular Mass Spectrometry
Protein Identification and PTM mapping
<h2 class="heading-underline">Protein Identification</h2>
<table cellpadding="0" cellspacing="0" style="width:500px;border: 0px solid white;margin-left:auto;margin-right:auto;" class="heading-underline"><tbody><tr><td style="border: 0px solid white"><img alt="protein ID and PTM determination" src="/biologicalsciences/images/proteinid.png" /></td><td style="text-align: left;vertical-align:text-top;border: 0px solid white"><strong>Useful for:</strong><ul><li>Confirming protein identity</li><li>Identifying unknown bands on SDS-PAGE gels</li><li>Detecting and localising PTMs</li><li>Determining interacting partners in pull downs</li></ul></td></tr></tbody></table>
Samples for protein identification should be submitted in the form of either a 1D gel that has been Coomassie stained or in a suitable buffer solution.
Please note that we cannot perform intact molecular mass measurements on samples supplied in gels, these must be prepared in solution or lyophilised.
Gel Samples
For gel samples, once the gel has been run, it should be handled inside a laminar flow hood where possible. This is to reduce contamination with keratin proteins.
Rinsing all boxes, eppendorfs, gel plates, scalpels etc. that come into contact with the gel with 70% ethanol can help to reduce the risk of keratin contamination.
Staining should be performed in clean gel boxes that have not been used for western blots.
Where possible Expedeon InstantBlue Comassie stain should be used as this is MS compatible.
Do not use excessively long staining times, bands should start to appear within 15 min depending upon the amount of material present.
The MS Facility can provide aliquots of Expedeon InstantBlue stain for internal users using the service for the first time.
If you require ID of bands detected using silver staining, please contact the Facility.
Internal users should provide the entire gel to the Facility in water along with a photograph indicating which bands to excise for analysis.
If you wish to submit a sample in solution please see below for the minimum sample amounts.
Ideally 1 – 5 ug protein should be loaded where possible.
The image below shows Coomassie-stained gel bands at different levels of protein loading:
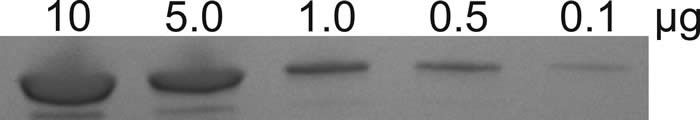
To be suitable for protein ID analysis, there must be a clearly defined band as shown for each loading level in the picture above.
Generally, the higher the loading on the gel the higher the sequence coverage achieved.
The band appearance at different loading levels will vary protein to protein.
Bands that are feint and/or diffuse, or require extreme settings during image acquisition to visualise are unlikely to give good sequencing results.
If we need to tilt or hold a gel up to the light to visualise a band, it is probably not suitable for analysis!
In-solution samples
We use S-Trap columns for in-solution samples. These provide a fast, reproducible and universal approach for digetsion and preparation of protein samples in solution.
In order to select the appropriate column to use, please provide a measure of total protein concentration with your submission. S-trap micro columns are used for sub-µg to 50 µg protein amounts and S-Trap minis are used for 50-300 µg of protein.
Almost all buffer components can be accepted, Urea (even 8M), salts, glycerol, PEG, other detergents, Ficoll, tween, triton, Lamelli loading buffer are fine. THe only exception is high concentrations of guanidinium chloride. Guanidinium concentration should be well below 6M concentration.
The ideal submission volume is ~25 µL, if the sample is much larger than this we will dry it down prior to analysis.
Sample submission deadlines
Protein ID is run on a weekly basis depending on demand.
Processing begins each Tuesday and bands should be submitted no later than midday on the Tuesday morning.
Any samples submitted after this time may be held over until the following week.
For new users, please come to the MS lab and have a chat with a member of the MS Facility staff and we can give you all the information that you need to get started.
Sample analysis can be requested through our online booking system iLab. Internal users can log in with their Univeristy credentials.
Please note that by submitting samples to the MS Facility for analysis you agree to our conditions of service and publications policy. A copy of which can be found here.
Users external to the University can find additional information here.
Notes
When submitting samples, you should provide the name of the species used to express the protein as well as the expected species of the expressed protein.
If the expected protein sequence differs from the wild type sequence in any way (e.g. inclusion of His tag or presence of TEV cleavage site) or the sequence is not present in the Uniprot database, please email the sequence in FASTA format to mass-spec@leeds.ac.uk.

